Chapter 13 Functional profiling
It is possible to investigate functional differences between metagenome (and metatranscriptome) samples by directly interrogating the read data. We will look at how this can be done with a package called HUMAnN (The HMP Unified Metabolic Analysis Network), a bioBakery pipeline designed to accurately profile the presence/absence and abundance of microbial pathways in metagenomic sequencing data.
HUMAnN is on its third version and was developed in tandem with the third version of MetaPhlAn, a computational tool for profiling the composition of microbial communities from metagenomic data.
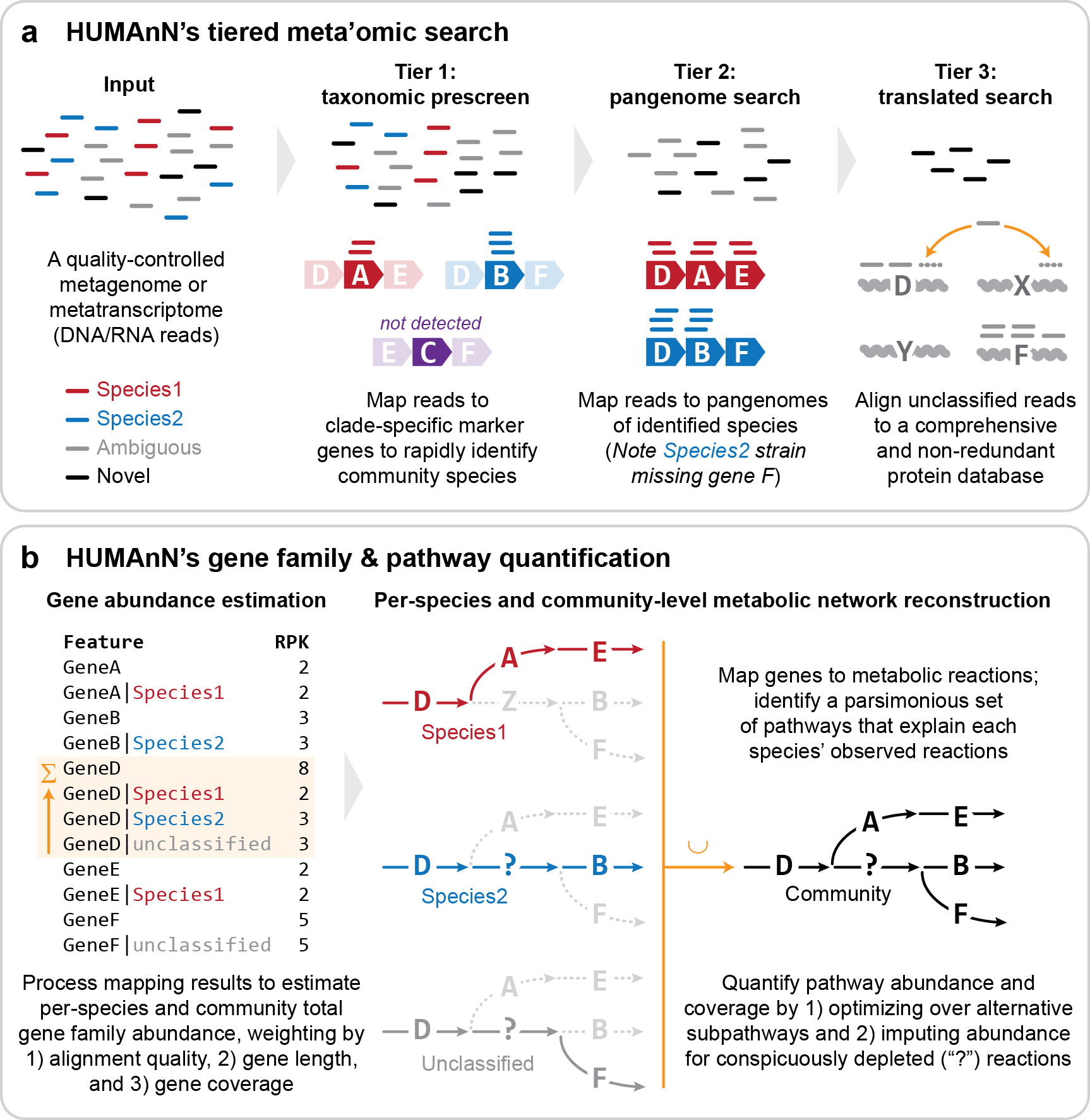
Please see below for a diagram showing the pipeline of HUMAnN:
HUMAnN 4 is currently in alpha: (https://forum.biobakery.org/t/announcing-humann-4-0-alpha/7531) I have tested it and could not install it correctly so we will not be teaching its use in this course till it is out of alpha and stable.